Visual Analysis of Time-Dependent Observables in Cell Signaling Simulations
Abstract
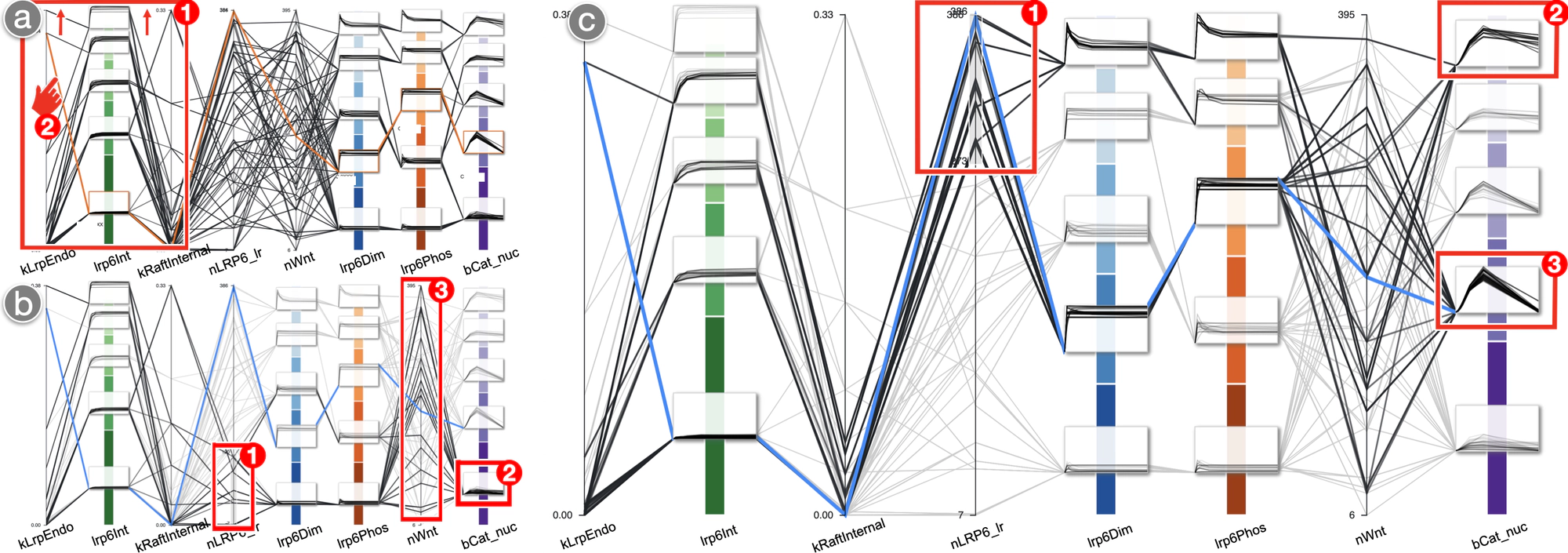
The ability of a cell to communicate with its environment is essential for key cellular functions like replication, differentiation, or metabolism. The involved molecular mechanisms are highly dynamic and difficult to capture experimentally. Simulation studies offer a valuable means for exploring and predicting how cell signaling processes unfold. We present a design study on the visual analysis of such studies to support 1) modelers in calibrating model parameters such that the simulated signal responses over time reflect reference behavior from cell biology research and 2) cell biologists in exploring the influence of receptor trafficking on the efficiency of signal transmission within the cell. We embed time series plots into parallel coordinates to enable a simultaneous analysis of model parameters and temporal outputs. A usage scenario illustrates how our approach assists with typical tasks such as assessing the plausibility of temporal outputs or examining their sensitivity across model configurations.
Type
Publication
Proceedings of the Eurographics Workshop on Visual Computing for Biology and Medicine - Short Papers